TRANSCRIPTION
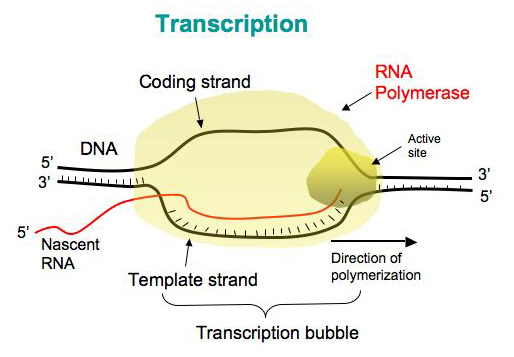
Transcription occurs in the nucleus and is when the DNA code from one strand of the DNA double helix is "rewritten" as a strand of RNA
Steps:
1. DNA strands separate to form a transcription bubble. One strand will be the template, the other will be "silent".
2. RNA polymerase binds to the template strand and produces a complementary RNA copy.
3. RNA separates from the template strand. It is called the primary RNA transcript.
4. Splicing: The primary RNA transcript is made up of coding (exon) and non-coding (intron) portions. The introns are cut out (spliced) by enzymes called spliceosomes and the exons are attached together.
5. The RNA leaves the nucleus for the cytoplasm. If it is mRNA it will go to the ribosomes. If it is rRNA it forms part of a ribosome. If it is tRNA, it will bind to the appropriate amino acid.
6. DNA double helix reforms.
https://www.onlinebiologynotes.com/wp-content/uploads/2017/06/transcription-1.jpg
TRANSLATION
Translation occurs at the ribosome. This is where the code on the strand of mRNA is converted into a chain of amino acids. A long chain is referred to as a polypeptide and this forms a protein. This conversion occurs via a specific coding mechanism, referred to as triplet codons.
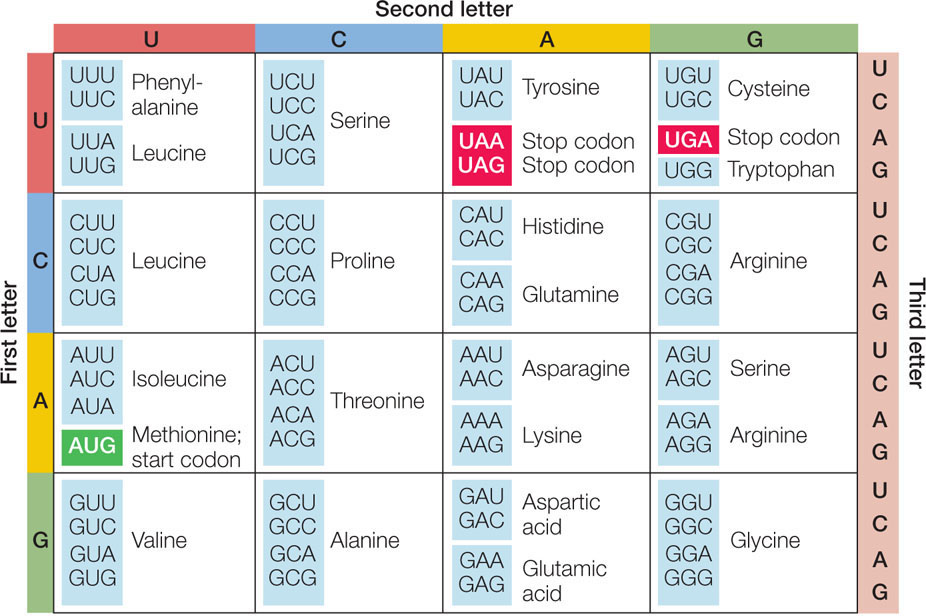
The mRNA is a long strand of nucleotides. Every 3 nucleotides form what is called a codon. They can be paired with three complementary nucleotides on the tRNA molecule. These are called the anticodon. The anticodon is found at one end of the tRNA. The other end of the tRNA is bound to a specific amino acid.
Scientists figured out which amino acid is attached to which type of tRNA and they have converted that information to form a codon table, which allows us to determine the order of amino acids if we know the nucleotide sequence found on a given mRNA.
https://www.macmillanhighered.com/BrainHoney/Resource/6716/digital_first_content/trunk/test/hillis2e/asset/img_ch10/c10_fig11.jpg
If you look at the table, you will note there are some codons that are highlighted. These are the start and stop codons, which indicate to the ribosome, where to begin and to end "reading" the mRNA. The start codon is always coded by the three nucleotide bases AUG (adenine, uracil, guanine). The complementary tRNA anticodon is UAC and it is bound to the amino acid methionine. Therefore, all polypeptide chains begin with the amino acid methionine. The stop codons do not code for any amino acids, they simply indicate to the ribosome that it should release the mRNA and the polypeptide chain.
This process occurs in a few steps:
1. Initiation: The mRNA binds to the ribosome and the first complementary tRNA binds with the mRNA. The tRNA is attached to a specific amino acid. All protein coding sequences begin with a "start codon".
2. Elongation: A new tRNA (complementary to the next codon) enters the ribosome and binds. If the wrong tRNA enters and cannot pair base pair with the mRNA, it is rejected. After the base pair binding occurs, the ribosome shifts one triplet along the mRNA and a new tRNA can enter and the procedure repeats. Since the amino acids attached to the tRNAs are in close contact with each other, they bind and begin to form a polypeptide chain. As the "oldest" tRNA shifts into the last position in the ribosome, it releases its amino acid, then leaves the ribosome. (It will go and find a free-floating amino acid in the cell cytoplasm to bind to, so that it is ready to take part in translation again).
3. Termination: When the ribosome reaches one of the stop codons (UGA, UAA, UAG) there will be no complementary tRNA to bind to. Instead, termination proteins (release factor) bind to the ribosome, causing it to dissociate and release the polypeptide chain. The ribosome can now pick up a new mRNA and begin the translation process again.
https://upload.wikimedia.org/wikipedia/commons/0/0f/Peptide_syn.png2. Elongation: A new tRNA (complementary to the next codon) enters the ribosome and binds. If the wrong tRNA enters and cannot pair base pair with the mRNA, it is rejected. After the base pair binding occurs, the ribosome shifts one triplet along the mRNA and a new tRNA can enter and the procedure repeats. Since the amino acids attached to the tRNAs are in close contact with each other, they bind and begin to form a polypeptide chain. As the "oldest" tRNA shifts into the last position in the ribosome, it releases its amino acid, then leaves the ribosome. (It will go and find a free-floating amino acid in the cell cytoplasm to bind to, so that it is ready to take part in translation again).
3. Termination: When the ribosome reaches one of the stop codons (UGA, UAA, UAG) there will be no complementary tRNA to bind to. Instead, termination proteins (release factor) bind to the ribosome, causing it to dissociate and release the polypeptide chain. The ribosome can now pick up a new mRNA and begin the translation process again.
DNA is made up of different types of genes:
1. Structural genes: code of proteins.
2. Regulatory genes: turn the structural genes on and off. Some genes don't need to be expressed all the time or throughout an individual's whole lifetime - for example, human haemoglobin has embryonic, fetal and adult forms, each of which are only expressed at the expressed at the appropriate time.
3. Modifier genes: these genes change the expression of other genes. For example, in the case of seasonal coat colour change, modifier genes are responsible for this.
4. Nonsense/filler: this DNA has no known function, although they may play a structural role in chromatin or chromosome folding.
These different kinds of DNA affect how and when a gene in the DNA will be transcribed and translated (or copied and decoded!). A few examples are given below, although regulation of expression is complex and still an area of much research.
Regulation in Prokaryotes
In prokaryotes, the DNA is organized in operons. An operon is the physical grouping of genes that act together, for example, the genes coding for proteins required in a biochemical pathway.
a. Induction: eg. E. coli's lac operon
If lactose is present (lac refers to the fact that the genes in the operon code for proteins involved in the breakdown of lactose), then the repressor protein binds the lactose molecules instead of binding to the DNA and then RNA polymerase can transcribe the DNA into mRNA. So, the presence of lactose induces transcription.
b. Repression: eg. E. coli's trp operon
The trp operon contains genes coding for proteins involved in tryptophan production. If tryptophan is not present, then the repressor protein is inactive, or doesn't bind to the DNA, so the RNA polymerase can bind and carry out transcription. In the presence of tryptophan, the repressor binds to the DNA, thus shutting down transcription.
https://i.ytimg.com/vi/6TgK8nhXXpA/maxresdefault.jpg
Regulation in eukaryotes is similar to that of prokaryotes (bacteria) in that it is primarily controlled at the level of transcription initiation and that it is controlled by proteins that bind to regulatory sequences, but it is more complex and sophisticated, with some of the following differences:
- eukaryotic genes are NOT clustered in operons, so each gene has its own regulatory elements.
- expression of genes can be affected by chromatin coiling and methylation.
- genes contain introns that are removed by RNA splicing. Splicing can remove different introns (or portions) of the DNA, resulting in the production of different mRNA, thus one stretch of DNA can, potentially, code for different proteins depending on how it is spliced.
Environmental control is far less common than in prokaryotes.
https://lh4.googleusercontent.com/Qr8ApKvaECqyD9dIC3xBA2D2rA998xCeuL06btyolWO38faRHaAH1coGWOxF-VchatRxyPf7dA2kpG_5oCIw-yfJiQZc8WYzvE9j4s7d118WugrVIIipHrWDlFdhIuVwwAUG



No comments:
Post a Comment